Natural Computing Lecture, March 15th, 2022
Artificial Immune Systems
Immunology's "negative selection" as anomaly detector
Inge Wortel (& Johannes Textor)
inge.wortel@ru.nl
Data Science, RU


Natural computing: the "bigger picture"
Two basic questions at the heart of this course:
- Can we design algorithms for computing science problems that are inspired by the natural world?
(i.e. to solve tasks such as information processing and optimization?) - Can we improve our understanding of the natural world by using computer science algorithms?
("What I cannot create, I do not understand" - Richard Feynman)
Previous two lectures: mostly [2]. Now: immuno-inspired computing — back to [1] (with a bit of [2]).
Why "immunology-inspired" computing?
The immune system has a number of attractive features worth replicating:
- layered protection (with multiple fail-safes)
- distributed protection (no single, "central" control to disable)
- (bio)diverse protection (no two persons are the same!)
- broad protection (even against previously unseen foreign material)
Why "immunology-inspired" computing?
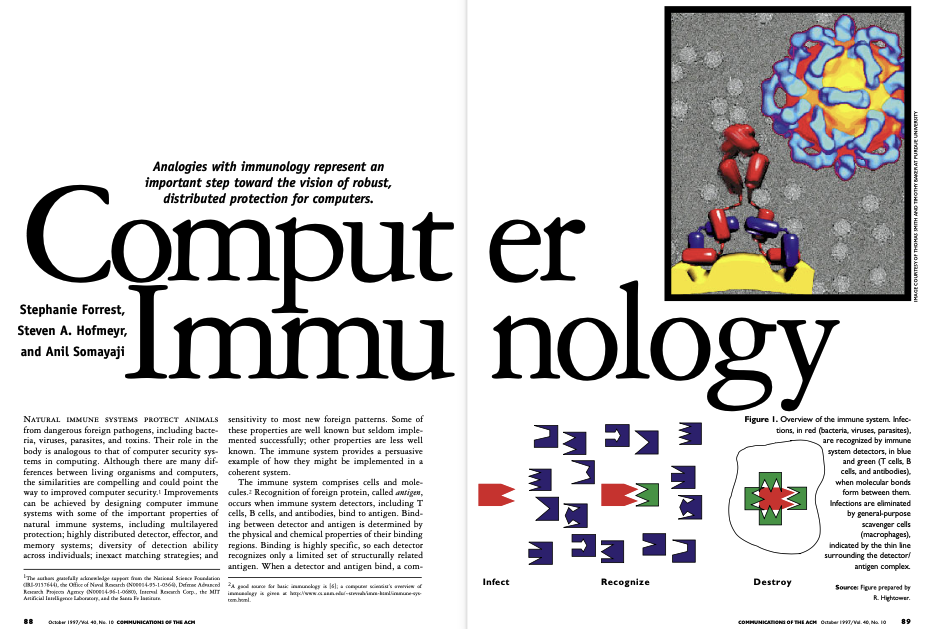
Stephanie Forrest, Steven A. Hofmeyr, and Anil Somayaji (1997). Communcations of the ACM, 40(10):88-96.
Artificial Immune Systems (AISs)
An artificial immune system is any algorithm/engineered system inspired by the real immune system.
The real immune system is incredibly complex and diverse — and so are AISs! But examples include:
- "Dendritic cell algorithm", inspired by a particular type of immune cell
- "Immune network algorithm", inspired by a theory of how the immune system works (now disproved)
- "B-cell algorithm", inspired by how the immune system makes antibodies
We will focus on one particular AIS: the negative selection algorithm.
Our source of inspiration: biological immune systems
Let's first examine how the immune system accomplishes these features:
- layered protection (with multiple fail-safes)
- distributed protection (no single, "central" control to disable)
- (bio)diverse protection (no two persons are the same!)
- broad protection (even against previously unseen foreign material)
| Question | |
|---|---|
| What do you think is the most important organ of our immune system? |
The immune system is multi-layered
Multiple layers of protection:
- The skin (otherwise: "mopping up the floor with the tap open")
- The complement system: generic & passive
- The innate immune system: cells that actively recognize (generic) pathogenic "patterns"
- The adaptive immune system: cells that actively combat specific pathogenic patterns
 |
 |
 |
 |
The (adaptive) immune system is distributed
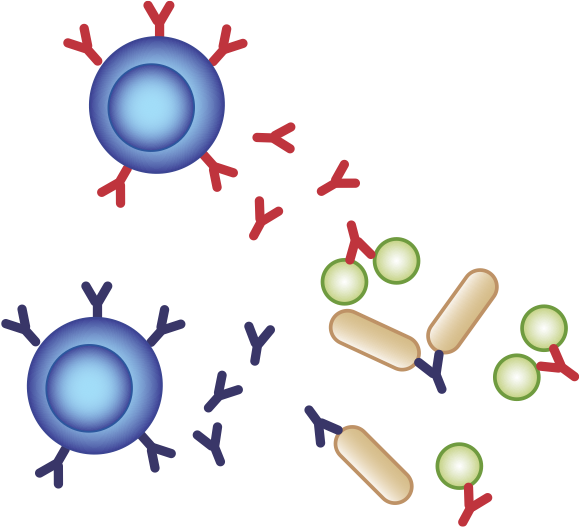
T cells
The adaptive immune system has two major subsystems: B cells and T cells
- Bursa (or bone marrow)-derived B cells make soluble antibodies to attack pathogens in the blood
- Thymus-derived T cells can attack and kill infected/cancerous cells
- Both are highly specialized or "specific":
we have many different cells that detect different things
T cells in action
| Question | |
|---|---|
| B cells make antibodies to attack pathogens in blood, whereas T cells attack pathogens inside our own cells. What do you think is harder — and why? |
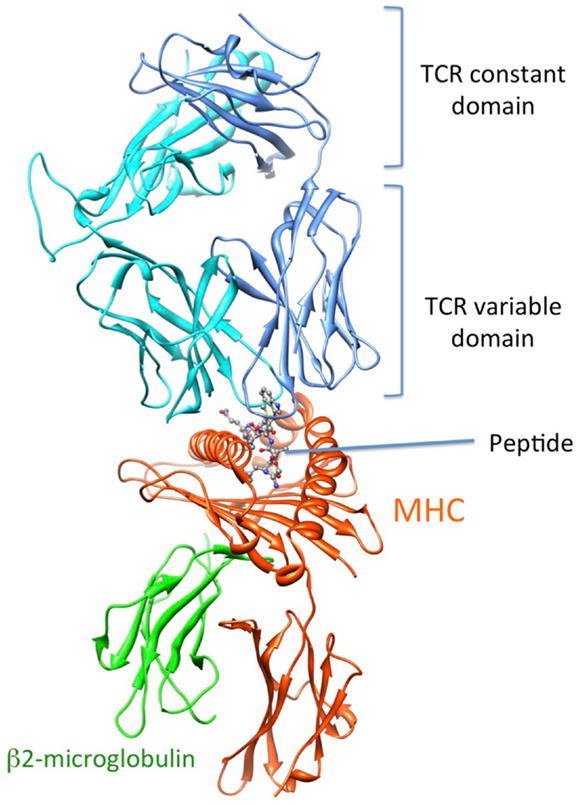
How T cells can see what is inside our cells
Human cells constantly make proteins and degrade them into fragments.
How T cells can see what is inside our cells
These fragments, peptides, are essentially strings of the amino acid "alphabet".
How T cells can see what is inside our cells
These fragments, peptides, are essentially strings of the amino acid "alphabet".
How T cells can see what is inside our cells
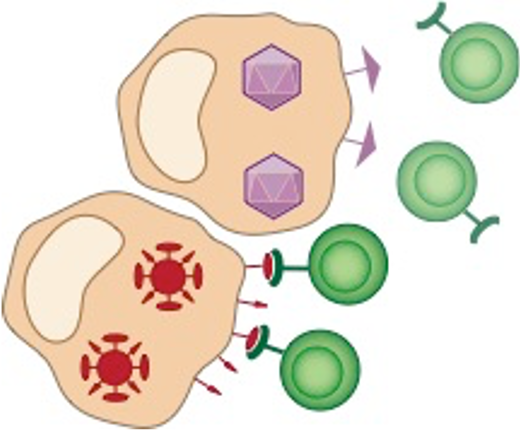
All cells (healthy or infected) "present" these peptides on their outer membrane, like little flags, using so-called MHC molecules.
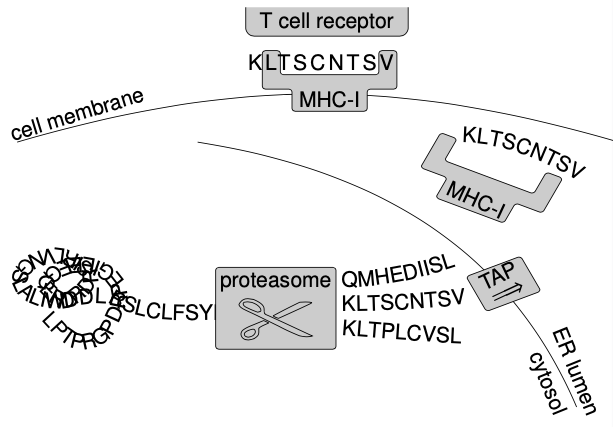
How T cells can see what is inside our cells
T cells constantly scan these MHC complexes for anomalous peptides:
- Healthy cells have only "normal" peptides derived from "normal" proteins
- Infected/cancerous/dying cells have "anomalous" peptides from foreign/mutated proteins
Their task becomes:
- kill infected/cancerous cells presenting (any!!!) anomalous peptides, while...
- ...tolerating healthy cells presenting normal peptides
The immune system is distributed and broad
T-cell immunity:
- A T cell binds to ("recognizes") a peptide using its T-cell receptor
- Each receptor is specific and binds to only ~ 1 in every 100 000 peptides
- Thus, we need many different receptors to recognize different pathogens
- To ensure this: T cells are diverse, with every T cell carrying its own receptor...
- ...such that protection is distributed over many (essentially unique) T cells
| Question | |
|---|---|
| Can you see a reason why this might be difficult? |
How do we get that many T cells?!
The problem:
- T-cell receptors are themselves proteins, encoded in our DNA
- We need (many) millions of T-cell receptors to "cover" all pathogens
- ... but our DNA only has space for ~20,000 genes...
How do we get that many T cells?!
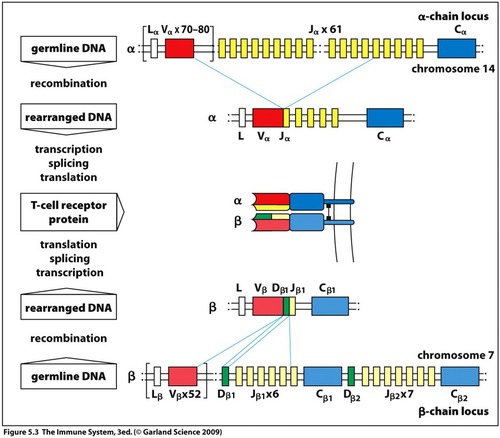
The problem:
- T-cell receptors are themselves proteins, encoded in our DNA
- We need (many) millions of T-cell receptors to "cover" all pathogens
- ... but our DNA only has space for ~20,000 genes...
The solution: randomize!
The immune system is diverse
Randomly generated T cells are diverse:
- within individuals
- between individuals
| Question | |
|---|---|
| Can you see both a benefit and a problem of this solution? |
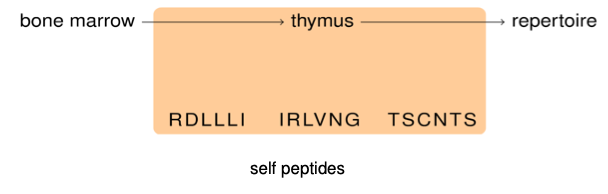
Negative selection
The (new) problem:
- Randomly generated T-cell receptors might detect also normal "self" peptides
- Such T cells can then attack our healthy cells; this is dangerous (auto-immunity)!
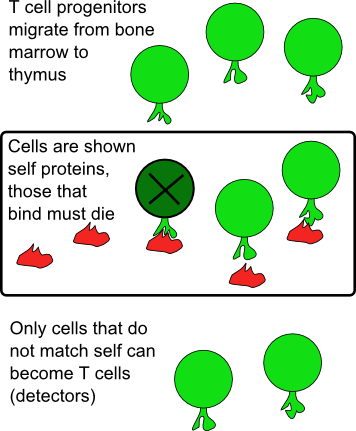
Solution — negative selection:
- T cells are born in the bone marrow, then go to the thymus for "training"
- There, they are then exposed to normal, "self" peptides...
- ..and they die if they recognize those.
- We are left with a T-cell repertoire where "self-reactive" cells have been deleted
Let's translate into ML language!
The immune system has a one-class classification problem
- We need to solve a binary classification problem ("normal" vs "anomalous" peptides)...
- ...but have only training examples from one class ("normal" peptides during negative selection)
Negative selection is an algorithm that solves this problem.
Negative selection algorithm: general structure
| Negative selection — IS |
|---|
 |
| Negative selection — algorithm | ||||
|---|---|---|---|---|
Let X be the learning domain containing both
for each m $\in$ M do (Forrest, 1997) |
Negative selection algorithm vs other population-based algorithms
| Discussion | |
|---|---|
|
Just like with other population-based algorithms (evolutionary algorithms, swarm intelligence, ...) we have a population of individuals (detectors) working together to solve a given task. But how is our AIS different in terms of:
|
Negative selection algorithm: one more thing
To make use of this generic scheme, we still need to define the detectors and the matching rule determining what they do and do not detect.
| Example 1: Real-valued negative selection | |
|---|---|
|
Learning domain: $\mathbb{R}^d$ (d-dim vectors) Detectors: hyperspheres, hypercubes, ... |
| Example 2: String-based negative selection | |
|---|---|
|
Learning domain: $\Sigma^\ell$ (strings of length $\ell$) Detectors: patterns |
Example 1: Real-valued negative selection (2D)
Learning domain X: $\mathbb{R}^2$
Detectors d: circles with fixed radius r
Generalization occurs, with a strength depending on r.
| Negative selection | ||||
|---|---|---|---|---|
|
| Matching rule |
|---|
|
Detector d matches any point s contained within its circle of radius r. |
Example 2: String-based negative selection
Learning domain X: $\{0,1\}^\ell$
Detectors d: patterns of length $\ell$ with parameter r
Generalization occurs, with a strength depending on r.
| Negative selection | ||||
|---|---|---|---|---|
|
| Matching rule: e.g. r-contiguous |
|---|
|
Detector d of length $\ell$ matches string s of length $\ell$ if d and s contain the same symbol at r subsequent positions. |
Using this to learn about biology
The (new) problem:
- Randomly generated T-cell receptors might detect also normal "self" peptides
- Such T cells can then attack our healthy cells; this is dangerous (auto-immunity)!
Solution — negative selection:
- T cells are born in the bone marrow, then go to the thymus for "training"
- There, they are then exposed to normal, "self" peptides...
- ..and they die if they recognize those.
- We are left with a T-cell repertoire where "self-reactive" cells have been deleted
Did we actually solve the problem?
- Wortel et al (2020). Cells. Is T Cell Negative Selection a Learning Algorithm?
- Blog: What if T cells could learn French?
Anomaly detection in Unix processes
Idea: trace unix system calls of a daemon process like sendmail, coding each system call as an alphabet symbol. Generate a database of system call sequences in normal circumstances.
Apply negative selection to these sequences and generate detectors, which may be capable to detect anomalous operations.
| Example | |
|---|---|
|
iKEKEEFEEgKEgKEEFEggFKvafvaegggggj 6LgggggngngnggLLnnPPggnnggLggiKEKE |
normal exploit |
So our strings don't have to be binary! In your assignment, you will apply negative selection to such data.
S. Forrest, S. A. Hofmeyr, A. Somayaji, T. A. Longstaff (1996). A sense of self for unix processes. IEEE Symposium on Security and Privacy.
How useful is such an AIS?
Back to our desirable features:
- layered protection (with multiple fail-safes)
- distributed protection (no single, "central" control to disable)
- (bio)diverse protection (no two persons are the same!)
- broad protection (even against previously unseen foreign material)
Our negative selection algorithm fulfils at least the last three. So why are they not more broadly used?
- This only works with large repertoires (/detector sets)
- ...implying large numbers of string comparisons, which has made too inefficient (so far).
- Improved implementations may solve this issue — but those are beyond the scope of this lecture.
Summary
The immune system's T cells are negatively selected to match anything but self.
A negative selection algorithm is any one-class classifier based on this principle, and can be applied to many data domains (but mostly strings).
Negative selection algorithms can learn from examples; i.e. they can generalize.
Negative selection requires a population ("repertoire") of detectors. But training does not yield a "fittest individual", but a fittest combination of detectors. This is important because every detector specializes in only a part of our learning domain X.
Implementing such an algorithm efficiently requires substantial work, but that is out of the scope of this lecture.
After the break, Gijs Schröder will talk about learning classifier systems, which have much in common with our AIS.