Eindhoven, May 23rd, 2022
What if T cells could learn French?
New answers to an old question in immunology
Inge Wortel
Computational Immunology group,
Radboud University Nijmegen, NL
inge.wortel@ru.nl![]()
@inge_wortel![]()
Should you take this bet?
- You get: 1 hour with my (French*) dictionary
- Test time: I show you words, you tell me if they are French (Y/N)
- You win: €10 per correct guess (but: you pay me €10 for every wrong one)
T cells recognize compromised cells specifically.
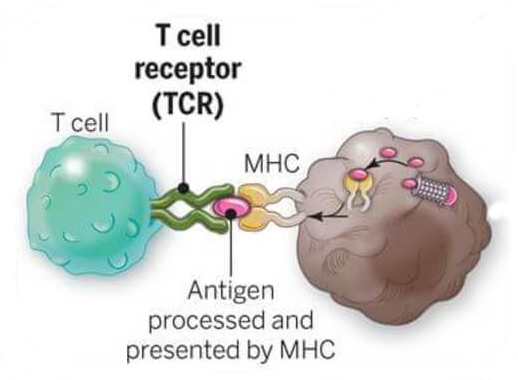
Adapted from National Cancer Institute (NIH)
- Adaptive immune system
- Recognize pathogen-infected or cancerous cells
- T-cell receptor (TCR) recognizes short peptides on MHC
- Compromised (infected/cancerous) cells display different peptides than healthy cells do
- Specific because each T cell's receptor recognises specific peptides
How many different peptides might your T cells encounter?
This number roughly equals the number of...
 |
 |
 |
| world-wide facebook users (109) | stars in our galaxy (1011) |
ants on earth (1015) |
T-cell diversity & tolerance: a problem of numbers
The immune system's T-cell repertoire must discriminate "self" vs "foreign":
- recognize >1015 foreign peptides ... 1
- ... while tolerating the many "self peptides" of healthy human cells


1 Sewell (2012). Nature Reviews Immunology.
Step 1: recognizing many foreign peptides
Make a repertoire with millions of T cells...
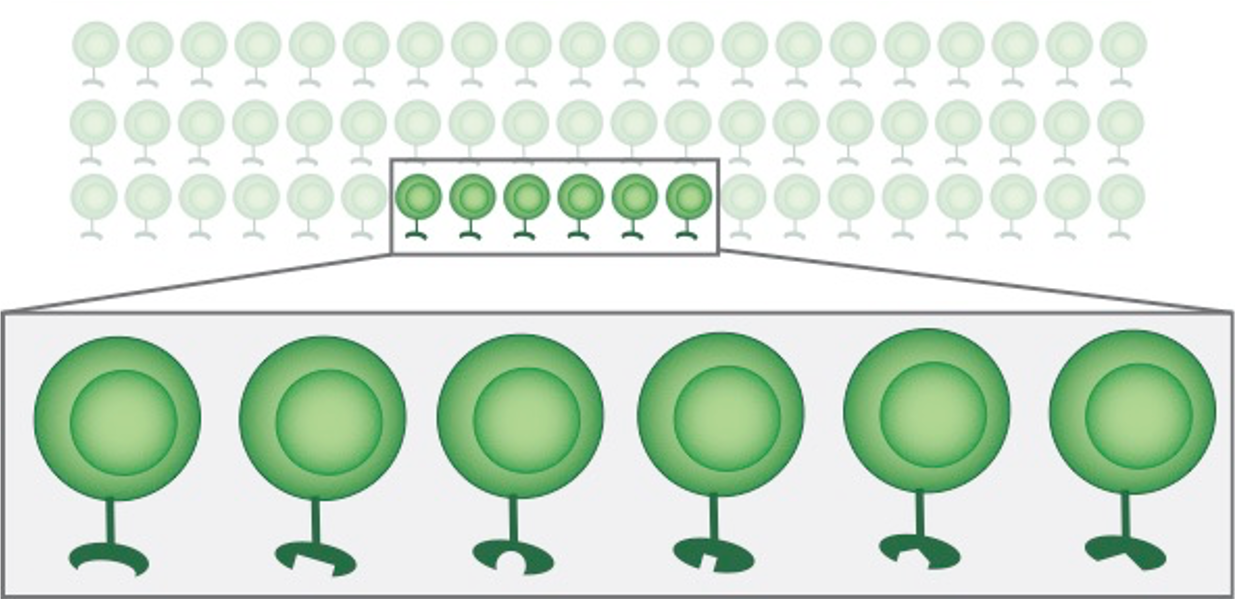
...each with a unique, specific receptor.
"Many hands make light work.""
Step 2: not recognizing self peptides
In the thymus, young T cells are "educated". Any cells that are self-reactive are filtered out: negative selection.
 |
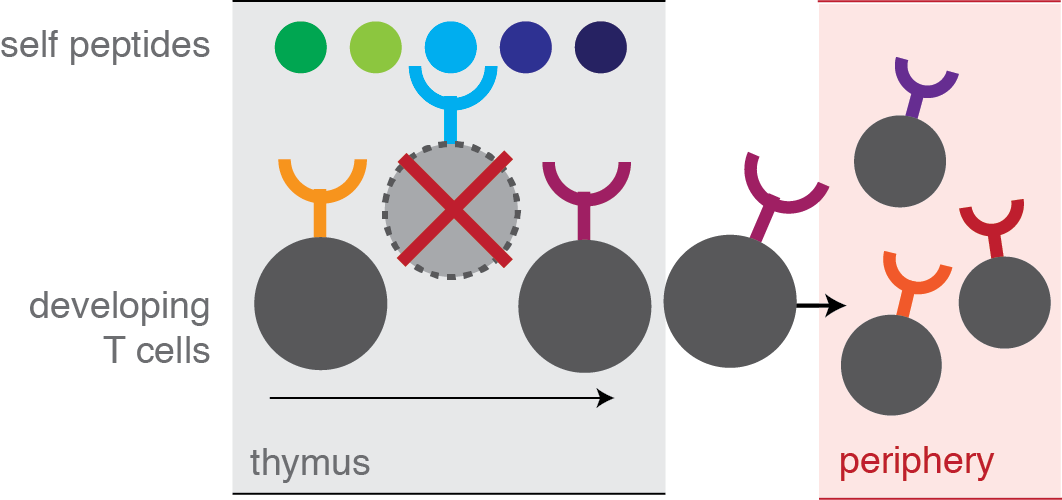 |
Sounds easy, but...
- T cells in the thymus have a few days to screen for half a million self-peptides...
- ... which roughly corresponds to 5 dictionaries!
- They can never see the entire "self dictionary"; negative selection is incomplete.

How "imperfect" is negative selection?
How different is the average recognition of a "self" peptide from that of a "foreign" peptide after negative selection?
- 3-fold
- 50-fold
- 100-fold
Sounds easy, but...
- T cells in the thymus have a few days to screen for half a million self-peptides...
- ... which roughly corresponds to 5 dictionaries!
- They can never see the entire "self dictionary"; negative selection is very incomplete.
So why do negative selection at all?

Let's vote
Are "tolerance" and "self-foreign discrimination" the same thing?
- Yes.
- No.
Tolerance ≠ discrimination.
Tolerance ≠ discrimination.
Which of these scenarios holds?
Given that negative selection is incomplete, can T cells distinguish
between self and foreign peptides they haven't seen
in the thymus?
Mission impossible?
Which word is French, and which is not?
- indoda
- fièvre
Goat cheese: learning by example

fièvre sounds/looks like chèvre

If we can learn which words look like French, can T cells in the thymus learn which peptides look like "self"?
Which of these scenarios holds?
Can T cells distinguish between self and foreign peptides they
haven't seen
in the thymus?
Which of these scenarios holds?
Can T cells learn by example during negative selection?
- in principle?
- in our immune system?
A simple model of TCR-sequence recognition
Three ingredients needed:
| 1 | Sequence | |
| 2 | TCR | |
| 3 | Affinity |
A simple model of TCR-sequence recognition
Three ingredients needed:
| 1 | Sequence | 6-letter strings of text in a certain language |
| 2 | TCR | |
| 3 | Affinity |
A simple model of TCR-sequence recognition
Three ingredients needed:
| 1 | Sequence | 6-letter strings of text in a certain language |
| 2 | TCR | binding motif |
| 3 | Affinity |
A simple model of TCR-sequence recognition
Three ingredients needed:
| 1 | Sequence | 6-letter strings of text in a certain language |
| 2 | TCR | binding motif |
| 3 | Affinity | Longest stretch of adjacent "matches" $\rightarrow$ binding if affinity $\geq$ threshold t |
Simulating negative selection in silico
Example: Xhosa recognition after negative selection on English
Negative selection allows discrimination
Negative selection on 500 English strings (t = 3),
Compare recognition of different English and Xhosa strings
| Motifs per string, before vs after: | Most frequently recognized: |
$\rightarrow$ Motifs distinguish strings they have not seen!
Why discrimination on unseen strings?
Motifs rarely react to both English and Xhosa:
Nodes:
- English
- Xhosa
Edge if >10,000 motifs in common
Concordance (same-language neighbors): 81%
Why discrimination on unseen strings?
This only works if strings are truly different:
Nodes:
- English
- More English
Edge if >10,000 motifs in common
Concordance (same-language neighbors): 50%
Two key requirements for discrimination
- Appropriate specificity/cross-reactivity
- Sufficient self-foreign dissimilarity
Two key requirements for discrimination
- Appropriate specificity/cross-reactivity
- Sufficient self-foreign dissimilarity
| Complete specificity (t = 6) | Low specificity (t = 1) | Intermediate specificity (t = 3) |
Two key requirements for discrimination
- Appropriate specificity/cross-reactivity
- Sufficient self-foreign dissimilarity
Summary: what have we learned from languages?
Proof of concept:
Negative selection can foster "learning by example".
- Self-foreign discrimination even on "unseen" sequences
- Even when tolerance is incomplete
But this works only if:
- self and foreign are sufficiently dissimilar
- cross-reactivity is intermediate
Back to self versus foreign peptides
Back to self versus foreign peptides
Use the same model for peptides instead of strings:
TCRs are moderately cross-reactive
Model at t = 4 matches order of magnitude of experimental estimates
| Model (t = 4) | Literature | |
| Cross-reactivity: | 1 : 55,000 peptides | 1 : 30,000 peptides 1 |
| Typical peptide recognition: | 0 - 20 TCRs / million | 0 - 100 TCRs / million 2-4 |
1 Ishizuka et al (2009). J Immunol.
2 Legoux et al (2010). J Immunol.
3 Blattman et al (2002). J Exp Med.
4 Alanio et al (2010). Blood.
How similar are "self" and "foreign" proteomes?
How similar are "self" and "foreign" proteomes?
Comparing "self" vs "foreign" peptides is like comparing English to:
- More English
- Medieval English
- Latin
- Xhosa
"self" and "foreign" peptides are similar
"self" and "foreign" peptides are similar
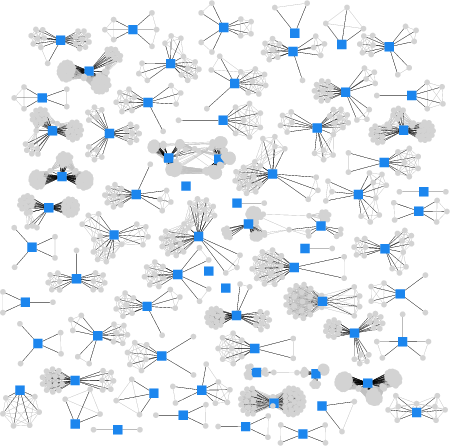
HIV peptides are embedded in
clusters of self peptides.
Xhosa/English: separate clusters.
$\rightarrow$ self-foreign discrimination will be difficult!
Self-foreign discrimination is difficult
... but remains possible:
What if thymic self peptides are non-random?
- random self peptides are often similar and will delete the same TCRs
$\rightarrow$ this is inefficient - some peptides are exchangeable, others are not
$\rightarrow$ a training set with more non-exchangeable peptides might do better:
"Optimal" training peptides improve self-foreign discrimination
Removal of self-reactivity $\neq$ self-foreign discrimination! Why does this work?
Why better self-foreign discrimination?
Self peptides with low exchangeability less often resemble foreign peptides:
$\rightarrow$ non-exchangeable peptides efficiently remove self-reactive TCRs, but preserve foreign-reactive ones!
How could the thymus be "optimal"?
Computed "optimal" set is enriched in rare AAs, depleted of common AAs.
Peptides with rare AAs tend to be less exchangeable:
$\rightarrow$ Could enrichment of rare AAs help self-foreign discrimination?
How could the thymus be "optimal"?
Choose "training" self peptides with a bias for peptides with rare AAs:
Simple bias already improves self-foreign discrimination.
The same is true for other pathogens
- Poor self-foreign discrimination after selection on random peptides
- Improved self-foreign discrimination after selection on biased peptides
Summary: self-foreign discrimination in the immune system

Negative selection allows "learning by example"
- Even incomplete negative selection can allow T cells
to discriminate "self" from "foreign" in principle. - This reconciles textbook theory with
surprising experimental data. - ...And it shows that our brain is not the only
organ that can learn!
...but self-foreign discrimination is hard:
- high self-foreign similarity hampers discrimination after selection
- Prediction: we can overcome this with training peptides biased for rare AAs
$\rightarrow$ because of AA bias in peptide presentation pathway?
Read more

Paper:
IMN Wortel, C Keşmir, RJ de Boer, JN Mandl, J Textor (2020). Is T-cell negative
selection a learning algorithm? Cells 9(3), 690; doi:10.3390/cells9030690
Blog post:
What if T cells could learn French? The Startup on Medium;
https://medium.com/swlh/what-if-t-cells-could-learn-french-8301f852254f?sk=c0dd70ee935cedefb8d35c670392d268
Acknowledgements
Tumor Immunology, Nijmegen
- Johannes Textor
- Jolanda de Vries
- Carl Figdor
- Human DLM
Funding
- Radboudumc PhD grant
- NWO
- KWF
- Horizon 2020
Theoretical Biology, Utrecht
- Rob de Boer
- Can Keşmir
Physiology, McGill University Montreal
- Judith Mandl



